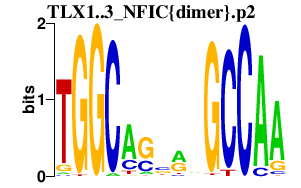
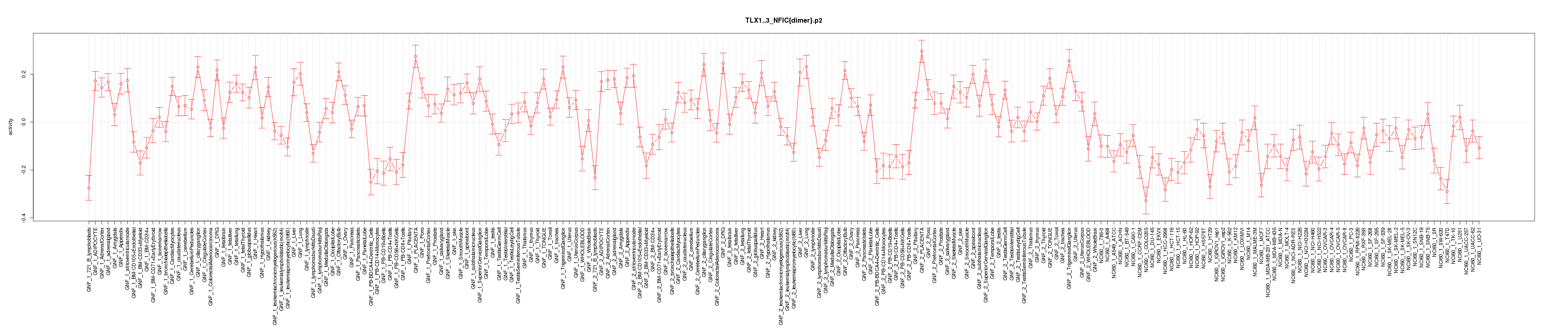
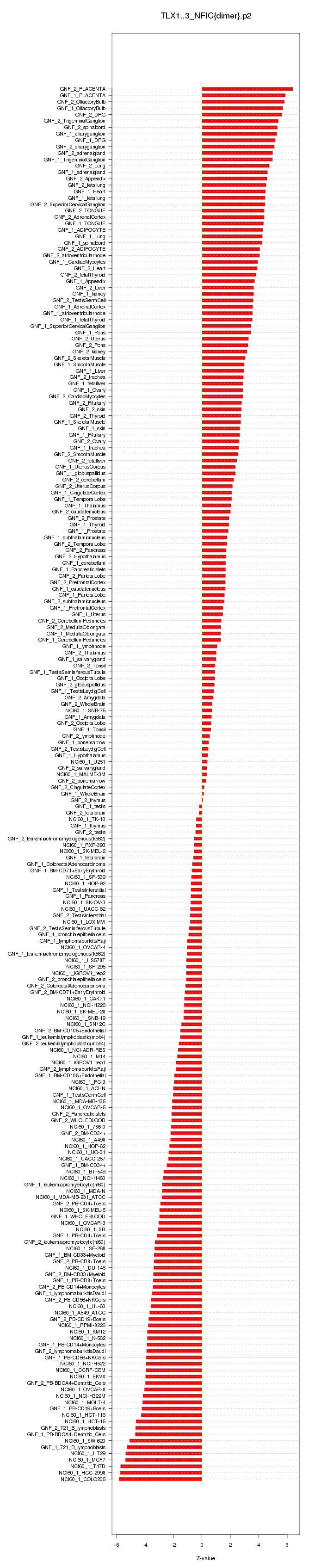
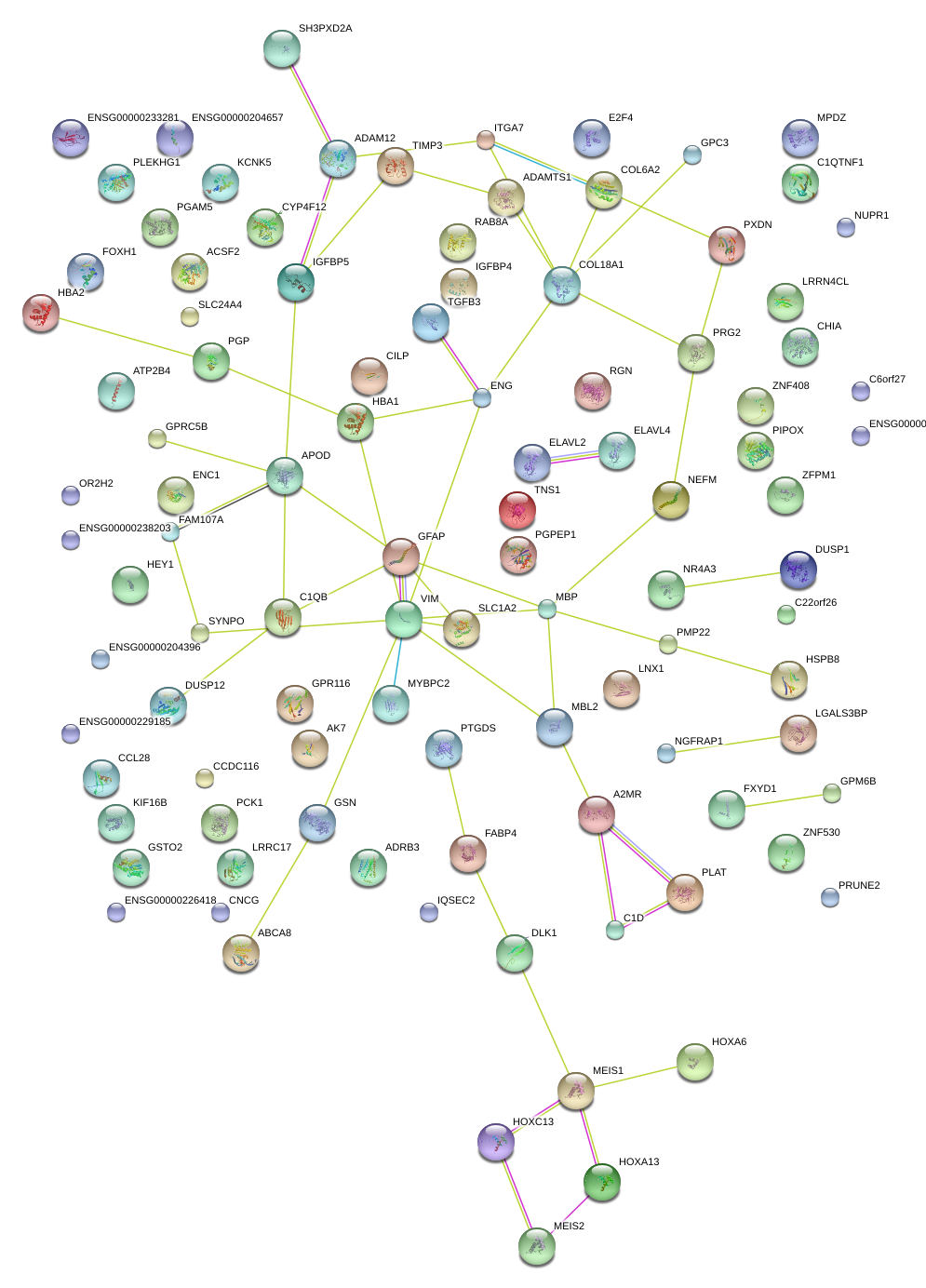
|
chr9_+_139872014
|
140.213
|
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr16_+_222845
|
109.580
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr16_+_226678
|
102.072
|
NM_000558
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chr9_+_139871954
|
99.060
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr18_-_74729049
|
71.449
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr18_-_74728963
|
69.730
|
|
MBP
|
myelin basic protein
|
|
chr2_-_218843541
|
60.699
|
|
TNS1
|
tensin 1
|
|
chr12_-_56106061
|
54.937
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr17_-_42992852
|
52.843
|
NM_001131019
NM_001242376
NM_002055
|
GFAP
|
glial fibrillary acidic protein
|
|
chrX_-_13956530
|
49.755
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_217560247
|
48.949
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr10_+_106034886
|
48.478
|
NM_001191014
NM_001191015
|
GSTO2
|
glutathione S-transferase omega 2
|
|
chrX_-_13956445
|
44.393
|
|
|
|
|
chr3_-_58563070
|
44.265
|
|
FAM107A
|
family with sequence similarity 107, member A
|
|
chr8_-_80679897
|
41.981
|
NM_001040708
NM_012258
|
HEY1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr6_+_150920998
|
41.750
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr17_-_66951381
|
38.848
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_-_195306274
|
38.128
|
|
APOD
|
apolipoprotein D
|
|
chr9_+_124086842
|
37.971
|
|
GSN
|
gelsolin
|
|
chr17_-_15164043
|
37.395
|
NM_153322
|
PMP22
|
peripheral myelin protein 22
|
|
chr21_+_46875402
|
34.560
|
NM_030582
|
COL18A1
|
collagen, type XVIII, alpha 1
|
|
chrX_-_13956644
|
33.651
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr16_-_2264776
|
33.404
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr7_-_27187367
|
33.175
|
NM_024014
|
HOXA6
|
homeobox A6
|
|
chr9_-_79520878
|
32.779
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr2_+_66662516
|
32.395
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr11_+_46722564
|
32.174
|
NM_001184751
|
ZNF408
|
zinc finger protein 408
|
|
chrX_-_133119475
|
32.024
|
|
GPC3
|
glypican 3
|
|
chr11_-_62457001
|
31.450
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr5_+_72251869
|
30.155
|
|
|
|
|
chr12_+_57522247
|
29.850
|
NM_002332
|
LRP1
|
low density lipoprotein receptor-related protein 1
|
|
chr9_-_23821808
|
29.612
|
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr10_-_128077062
|
29.590
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr10_-_105452878
|
29.559
|
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chrX_+_46937807
|
29.484
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr10_+_17271302
|
28.949
|
|
VIM
|
vimentin
|
|
chr6_-_46922533
|
28.553
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr17_+_27369917
|
28.267
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chrX_+_46937889
|
28.246
|
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chr12_+_119616594
|
28.121
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chrX_+_46937744
|
28.108
|
NM_004683
NM_152869
|
RGN
|
regucalcin (senescence marker protein-30)
|
|
chrX_-_133119624
|
28.044
|
NM_001164617
NM_001164618
NM_001164619
NM_004484
|
GPC3
|
glypican 3
|
|
chr10_+_17271277
|
28.036
|
|
VIM
|
vimentin
|
|
chr5_+_150020201
|
28.030
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr9_-_13279562
|
28.010
|
|
MPDZ
|
multiple PDZ domain protein
|
|
chrX_-_53310748
|
27.593
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr5_-_172198160
|
27.575
|
NM_004417
|
DUSP1
|
dual specificity phosphatase 1
|
|
chr19_+_15783827
|
27.515
|
NM_023944
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12
|
|
chr15_-_65503786
|
27.195
|
NM_003613
|
CILP
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase
|
|
chr9_-_23821842
|
27.147
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_+_66662692
|
26.883
|
|
MEIS1
|
Meis homeobox 1
|
|
chr21_-_28217272
|
26.863
|
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr19_+_58111244
|
26.819
|
NM_020880
|
ZNF530
|
zinc finger protein 530
|
|
chr7_+_102553343
|
26.439
|
NM_001031692
NM_005824
|
LRRC17
|
leucine rich repeat containing 17
|
|
chr1_+_22979681
|
26.346
|
NM_000491
|
C1QB
|
complement component 1, q subcomponent, B chain
|
|
chr9_-_130616957
|
25.945
|
|
ENG
|
endoglin
|
|
chr17_+_38599651
|
25.804
|
NM_001552
|
IGFBP4
|
insulin-like growth factor binding protein 4
|
|
chr6_+_29555682
|
25.579
|
NM_007160
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2
|
|
chr10_+_17271341
|
25.579
|
|
VIM
|
vimentin
|
|
chr6_-_31745048
|
25.119
|
NM_025258
|
C6orf27
|
chromosome 6 open reading frame 27
|
|
chr11_-_62457370
|
24.694
|
NM_203422
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr10_+_17271276
|
24.623
|
|
VIM
|
vimentin
|
|
chr3_-_14989399
|
24.409
|
|
LOC100505641
|
uncharacterized LOC100505641
|
|
chr9_-_130616592
|
24.286
|
|
ENG
|
endoglin
|
|
chr14_-_76448091
|
24.079
|
NM_003239
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr22_+_21986968
|
23.970
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr4_-_48014960
|
23.936
|
NM_000087
|
CNGA1
|
cyclic nucleotide gated channel alpha 1
|
|
chr17_+_27370155
|
23.545
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr22_+_33196801
|
23.523
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr8_-_82395401
|
23.447
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr21_-_28217692
|
23.347
|
NM_006988
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1
|
|
chr19_+_16222706
|
23.096
|
|
RAB8A
|
RAB8A, member RAS oncogene family
|
|
chr9_+_124043719
|
22.869
|
|
GSN
|
gelsolin
|
|
chr21_+_47531369
|
22.825
|
|
COL6A2
|
collagen, type VI, alpha 2
|
|
chr12_+_54332575
|
22.660
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr19_+_50936159
|
22.446
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr8_-_42065193
|
22.264
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr9_+_102584136
|
22.126
|
NM_006981
NM_173199
NM_173200
|
NR4A3
|
nuclear receptor subfamily 4, group A, member 3
|
|
chr8_+_24772454
|
22.024
|
NM_001105541
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr9_-_130616760
|
21.731
|
|
ENG
|
endoglin
|
|
chr20_+_56136136
|
21.618
|
NM_002591
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble)
|
|
chr8_-_37822813
|
21.466
|
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr14_+_92789536
|
21.384
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr19_+_35630391
|
21.366
|
NM_005031
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chr17_+_77030471
|
21.299
|
NM_198593
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1
|
|
chr14_+_101193275
|
21.188
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr11_-_35440514
|
21.156
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr16_-_19896105
|
20.641
|
NM_016235
|
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B
|
|
chr5_-_73937248
|
20.484
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr6_-_39197225
|
20.412
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr1_+_203595929
|
20.348
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr1_+_111833473
|
20.229
|
NM_021797
NM_201653
|
CHIA
|
chitinase, acidic
|
|
chr8_-_145701717
|
20.031
|
NM_003923
|
FOXH1
|
forkhead box H1
|
|
chrX_+_102631408
|
20.029
|
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr17_-_76975932
|
19.968
|
NM_005567
|
LGALS3BP
|
lectin, galactoside-binding, soluble, 3 binding protein
|
|
chr14_+_96858436
|
19.950
|
NM_152327
|
AK7
|
adenylate kinase 7
|
|
chr17_+_48503607
|
19.787
|
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr16_+_67226027
|
19.761
|
NM_001950
|
E2F4
|
E2F transcription factor 4, p107/p130-binding
|
|
chr22_-_46449972
|
19.755
|
NM_018280
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr7_-_64023444
|
19.700
|
|
|
|
|
chrX_+_102631267
|
19.686
|
NM_206915
NM_206917
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr5_-_43412417
|
19.617
|
NM_148672
|
CCL28
|
chemokine (C-C motif) ligand 28
|
|
chr16_-_28550324
|
19.487
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr14_+_92788924
|
19.367
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr8_-_42065037
|
19.281
|
|
PLAT
|
plasminogen activator, tissue
|
|
chr3_-_12200850
|
19.239
|
NM_003256
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|
|
chr1_+_153600859
|
19.142
|
NM_006271
|
S100A1
|
S100 calcium binding protein A1
|
|
chr14_+_101193313
|
19.051
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr1_+_203596222
|
18.987
|
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr1_+_203595897
|
18.977
|
NM_001001396
NM_001684
|
ATP2B4
|
ATPase, Ca++ transporting, plasma membrane 4
|
|
chr6_+_31982538
|
18.746
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr10_-_128077011
|
18.680
|
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr14_-_76447445
|
18.671
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr1_-_23810627
|
18.476
|
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr3_+_14989076
|
18.364
|
NM_003298
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr22_+_22735202
|
18.349
|
|
|
|
|
chr1_-_243418675
|
18.047
|
NM_001042404
NM_001042405
NM_014812
|
CEP170
|
centrosomal protein 170kDa
|
|
chr16_+_2588000
|
18.045
|
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr10_+_88718287
|
17.878
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr22_+_26565623
|
17.818
|
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr9_-_113800243
|
17.756
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr22_+_30792929
|
17.615
|
NM_001204204
NM_012429
NM_033382
|
SEC14L2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr9_-_34710062
|
17.516
|
NM_002989
|
CCL21
|
chemokine (C-C motif) ligand 21
|
|
chr15_-_40213087
|
17.436
|
NM_007223
|
GPR176
|
G protein-coupled receptor 176
|
|
chr1_+_154377847
|
17.374
|
|
IL6R
|
interleukin 6 receptor
|
|
chr19_+_46806855
|
17.308
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr10_+_124320180
|
17.128
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr17_+_77893155
|
17.060
|
|
|
|
|
chr22_+_41763299
|
16.956
|
NM_001145398
|
TEF
|
thyrotrophic embryonic factor
|
|
chr1_-_22263676
|
16.947
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr1_+_150255228
|
16.934
|
NM_144697
|
C1orf51
|
chromosome 1 open reading frame 51
|
|
chr9_+_27109138
|
16.496
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr1_+_1981902
|
16.425
|
NM_002744
|
PRKCZ
|
protein kinase C, zeta
|
|
chr9_+_124030455
|
16.290
|
|
GSN
|
gelsolin
|
|
chr6_+_31949800
|
16.245
|
NM_001242823
NM_001252204
NM_007293
NM_001002029
|
LOC100293534
C4A
C4B
|
complement C4-B-like
complement component 4A (Rodgers blood group)
complement component 4B (Chido blood group)
|
|
chr8_+_55370494
|
16.206
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr17_+_48503518
|
16.056
|
NM_025149
|
ACSF2
|
acyl-CoA synthetase family member 2
|
|
chr6_+_39760772
|
15.953
|
NM_015345
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr10_+_17270469
|
15.865
|
|
VIM
|
vimentin
|
|
chr19_+_46801632
|
15.832
|
NM_022462
NM_152796
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr14_+_69951396
|
15.828
|
NM_001161498
|
PLEKHD1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1
|
|
chr14_+_101193190
|
15.730
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chrX_-_40506666
|
15.702
|
|
CXorf38
|
chromosome X open reading frame 38
|
|
chr9_-_130617009
|
15.688
|
NM_000118
NM_001114753
|
ENG
|
endoglin
|
|
chr1_-_201342318
|
15.655
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr22_-_19109718
|
15.576
|
|
DGCR2
|
DiGeorge syndrome critical region gene 2
|
|
chrX_-_154114431
|
15.555
|
NM_019863
|
F8
|
coagulation factor VIII, procoagulant component
|
|
chr6_+_132129195
|
15.534
|
|
ENPP1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1
|
|
chr7_+_116165089
|
15.501
|
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr11_+_114166534
|
15.296
|
NM_006169
|
NNMT
|
nicotinamide N-methyltransferase
|
|
chr19_+_50380681
|
15.280
|
NM_001168222
NM_024682
|
TBC1D17
|
TBC1 domain family, member 17
|
|
chr1_+_54359859
|
15.261
|
NM_000792
NM_001039715
NM_001039716
NM_213593
|
DIO1
|
deiodinase, iodothyronine, type I
|
|
chr11_-_6341708
|
15.202
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr14_+_85996454
|
15.002
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr17_+_7184978
|
14.906
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr19_+_58790306
|
14.716
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr1_-_16344438
|
14.706
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr2_-_216300444
|
14.617
|
|
FN1
|
fibronectin 1
|
|
chr3_-_42305438
|
14.612
|
NM_001174138
|
CCK
|
cholecystokinin
|
|
chr16_-_86542463
|
14.568
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr20_+_42194736
|
14.355
|
NM_016276
|
SGK2
|
serum/glucocorticoid regulated kinase 2
|
|
chr11_-_2159472
|
14.199
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_173174611
|
14.174
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr19_+_55281262
|
14.154
|
NM_014218
|
KIR2DL1
KIR2DS4
|
killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 1
killer cell immunoglobulin-like receptor, two domains, short cytoplasmic tail, 4
|
|
chr7_-_97619350
|
14.106
|
NM_006188
|
OCM2
|
oncomodulin 2
|
|
chr1_-_32264215
|
14.019
|
|
SPOCD1
|
SPOC domain containing 1
|
|
chr8_+_38243958
|
14.000
|
NM_001199659
NM_144652
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2
|
|
chr7_-_44105162
|
13.965
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr10_+_17271859
|
13.942
|
|
VIM
|
vimentin
|
|
chr6_-_30043547
|
13.836
|
NM_170769
NM_025236
|
RNF39
|
ring finger protein 39
|
|
chr20_+_34742656
|
13.817
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr15_-_35087748
|
13.775
|
NM_005159
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr15_+_42867708
|
13.743
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr19_-_14016908
|
13.610
|
NM_024323
|
C19orf57
|
chromosome 19 open reading frame 57
|
|
chr6_+_14117486
|
13.524
|
NM_001251901
|
CD83
|
CD83 molecule
|
|
chr21_+_27107322
|
13.503
|
NM_002040
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa
|
|
chr19_-_41356193
|
13.239
|
NM_000762
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6
|
|
chr3_+_9791602
|
13.210
|
NM_002542
NM_016819
NM_016820
NM_016821
NM_016826
NM_016827
NM_016828
NM_016829
|
OGG1
|
8-oxoguanine DNA glycosylase
|
|
chr1_+_154378044
|
13.205
|
|
IL6R
|
interleukin 6 receptor
|
|
chr1_-_112046742
|
13.198
|
NM_000677
NM_020683
|
ADORA3
|
adenosine A3 receptor
|
|
chr2_+_203499819
|
13.193
|
NM_173511
|
FAM117B
|
family with sequence similarity 117, member B
|
|
chr10_-_128077072
|
13.190
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr7_-_5569200
|
13.153
|
|
ACTB
|
actin, beta
|
|
chr1_-_23810721
|
13.147
|
NM_001143778
NM_017707
|
ASAP3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3
|
|
chr3_-_126076057
|
13.088
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr8_+_17780492
|
13.070
|
|
PCM1
|
pericentriolar material 1
|
|
chr13_+_113622758
|
13.066
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr15_-_90347173
|
13.049
|
|
ANPEP
|
alanyl (membrane) aminopeptidase
|
|
chr11_+_1244292
|
13.044
|
NM_002458
|
MUC5B
|
mucin 5B, oligomeric mucus/gel-forming
|
|
chr16_+_20420854
|
13.036
|
NM_017888
|
ACSM5
|
acyl-CoA synthetase medium-chain family member 5
|
|
chr20_+_24450269
|
13.000
|
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr8_+_22022644
|
12.993
|
NM_001199
NM_006129
|
BMP1
|
bone morphogenetic protein 1
|
|
chr8_+_22022677
|
12.959
|
|
BMP1
|
bone morphogenetic protein 1
|
|
chr16_+_2587951
|
12.933
|
NM_002613
NM_031268
|
PDPK1
|
3-phosphoinositide dependent protein kinase-1
|
|
chr17_-_15902907
|
12.929
|
NM_001042697
NM_001042698
|
ZSWIM7
|
zinc finger, SWIM-type containing 7
|
|
chr14_+_95047792
|
12.854
|
|
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5
|
|
chr1_-_151345156
|
12.853
|
NM_003944
|
SELENBP1
|
selenium binding protein 1
|
|
chr19_-_40440532
|
12.840
|
NM_003890
|
FCGBP
|
Fc fragment of IgG binding protein
|
|
chr19_-_33716609
|
12.797
|
NM_019849
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10
|
|
chr3_-_12200339
|
12.670
|
|
TIMP4
|
TIMP metallopeptidase inhibitor 4
|